Difference between revisions of "Analysis Package"
(→Automated Segmentation) |
|||
| Line 35: | Line 35: | ||
<br clear=all> | <br clear=all> | ||
| + | == dF/F Trace Extraction and Deconvolution == | ||
== Alignment Across Sessions == | == Alignment Across Sessions == | ||
Revision as of 23:16, 13 January 2016
Contents
Overview
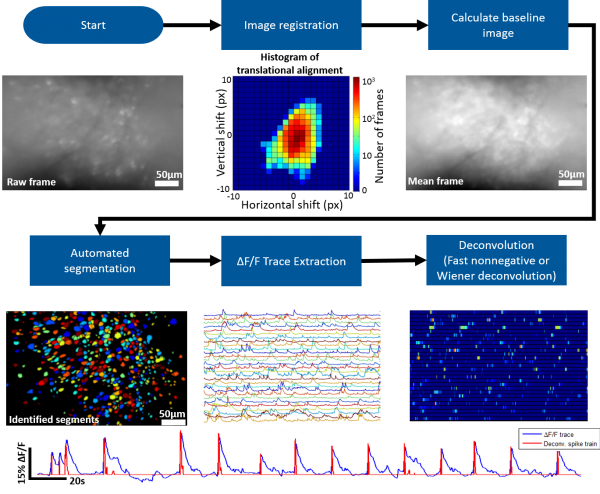
The MATLAB analysis package provided on this wiki contains the tools necessary for going from the raw microscope video to deconvolved individual neuron activity. Our package supports batch (generally run in parallel) processing for handling multiple recording sessions and can also be run on an individual session stored in the MATLAB workspace.
Roughly speaking the steps involved are the following
- Generate MATLAB data structure
- All access to the raw or processed data is handled through this data structure
- Memory efficient (less than 10MB per minute of recording)
- Correct for motion artifacts using an amplitude or FFT image registration algorithm
- Identify the bountries of individual neurons (we provide a novel, fully automated segmentation approach)
- Extract dF/F activity for identified neurons
- Deconvolve dF/F activity using Wiener or Fast Nonnegative deconvolution to approximate neuronal spiking pattern
- Additional Analysis:
- Cross-talk removal
- Segment matching across sessions
- Behavior tracking and syncing to microscope data
- Activity detection
The Miniscope Analysis Github repository can be found here.
Work Flow
A general work flow for processing and analyzing miniscope data can be found in msBatchRun.m. This script will walk you through all the necessary functions to go from the raw data to approximate spiking activity of individual neurons.
Image Registration
We suggest using Dario Ringach's recursive image registration algorithm.
Automated Segmentation